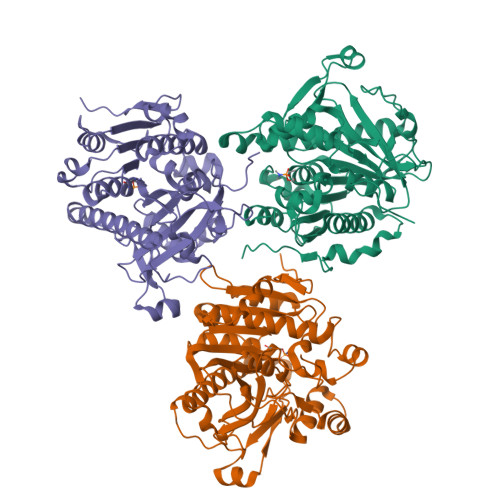
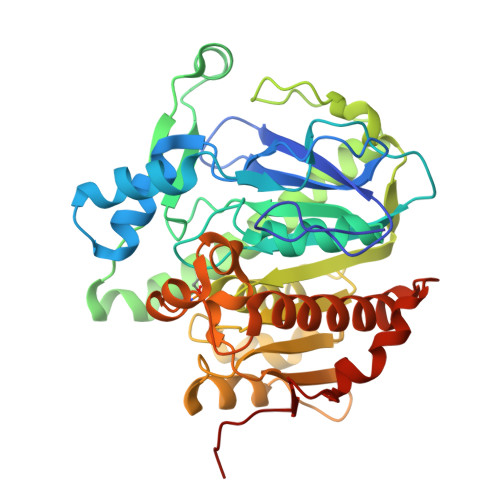
Crystal structures of human group-VIIA phospholipase A2 inhibited by organophosphorus nerve agents exhibit non-aged complexes.
Samanta, U., Kirby, S.D., Srinivasan, P., Cerasoli, D.M., Bahnson, B.J.(2009) Biochem Pharmacol 78: 420-429
- PubMed: 19394314
- DOI: https://doi.org/10.1016/j.bcp.2009.04.018
- Primary Citation of Related Structures:
3F96, 3F97, 3F98, 3F9C - PubMed Abstract:
The enzyme group-VIIA phospholipase A2 (gVIIA-PLA2) is bound to lipoproteins in human blood and hydrolyzes the ester bond at the sn-2 position of phospholipid substrates with a short sn-2 chain. The enzyme belongs to a serine hydrolase superfamily of enzymes, which react with organophosphorus (OP) nerve agents. OPs ultimately exert their toxicity by inhibiting human acetycholinesterase at nerve synapses, but may additionally have detrimental effects through inhibition of other serine hydrolases. We have solved the crystal structures of gVIIA-PLA2 following inhibition with the OPs diisopropylfluorophosphate, sarin, soman and tabun. The sarin and soman complexes displayed a racemic mix of P(R) and P(S) stereoisomers at the P-chiral center. The tabun complex displayed only the P(R) stereoisomer in the crystal. In all cases, the crystal structures contained intact OP adducts that had not aged. Aging refers to a secondary process OP complexes can go through, which dealkylates the nerve agent adduct and results in a form that is highly resistant to either spontaneous or oxime-mediated reactivation. Non-aged OP complexes of the enzyme were corroborated by trypsin digest and matrix-assisted laser desorption ionization mass spectrometry of OP-enzyme complexes. The lack of stereoselectivity of sarin reaction was confirmed by gas chromatography/mass spectrometry using a chiral column to separate and quantitate the unbound stereoisomers of sarin following incubation with enzyme. The structural details and characterization of nascent reactivity of several toxic nerve agents is discussed with a long-term goal of developing gVIIA-PLA2 as a catalytic bioscavenger of OP nerve agents.
Organizational Affiliation:
Department of Chemistry & Biochemistry, University of Delaware, Newark, DE 19716, USA.